华大空转Stereo-seq
Stereo-seq介绍 | Stereo-seq的技术优势 | 嘉因生物Stereo-seq服务优势
▍Stereo-seq介绍
空间转录组(Spatial Transcriptomics) 是一种用于研究细胞在组织切片上空间分布的实验技术。 传统上,转录组学主要关注基因在整体水平上的表达情况;空间转录组技术允许在组织切片上的成百上千个位置同时检测基因表达量,从而绘制出基因表达/细胞类型的空间图谱。
Stereo-seq 是华大研究院开发的空间转录组技术平台。该平台具有纳米级分辨率,厘米级全景视场两大特点。
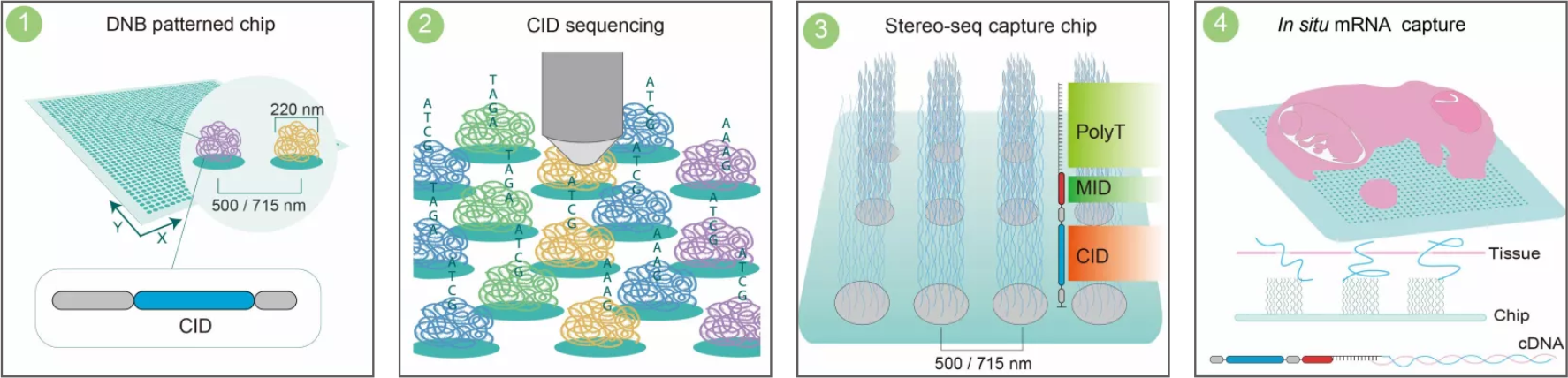
2023年2月,华大时空组学授权球盟会(中国)官方网站(以下简称嘉因生物)开展时空组学Stereo-seq实验,嘉因生物正式成为华大时空组学生态合作伙伴。

▍Stereo-seq的技术优势
优势一: 纳米级分辨率,实现细胞级分子定位。20*20个spots 覆盖一个直径为10um的细胞。
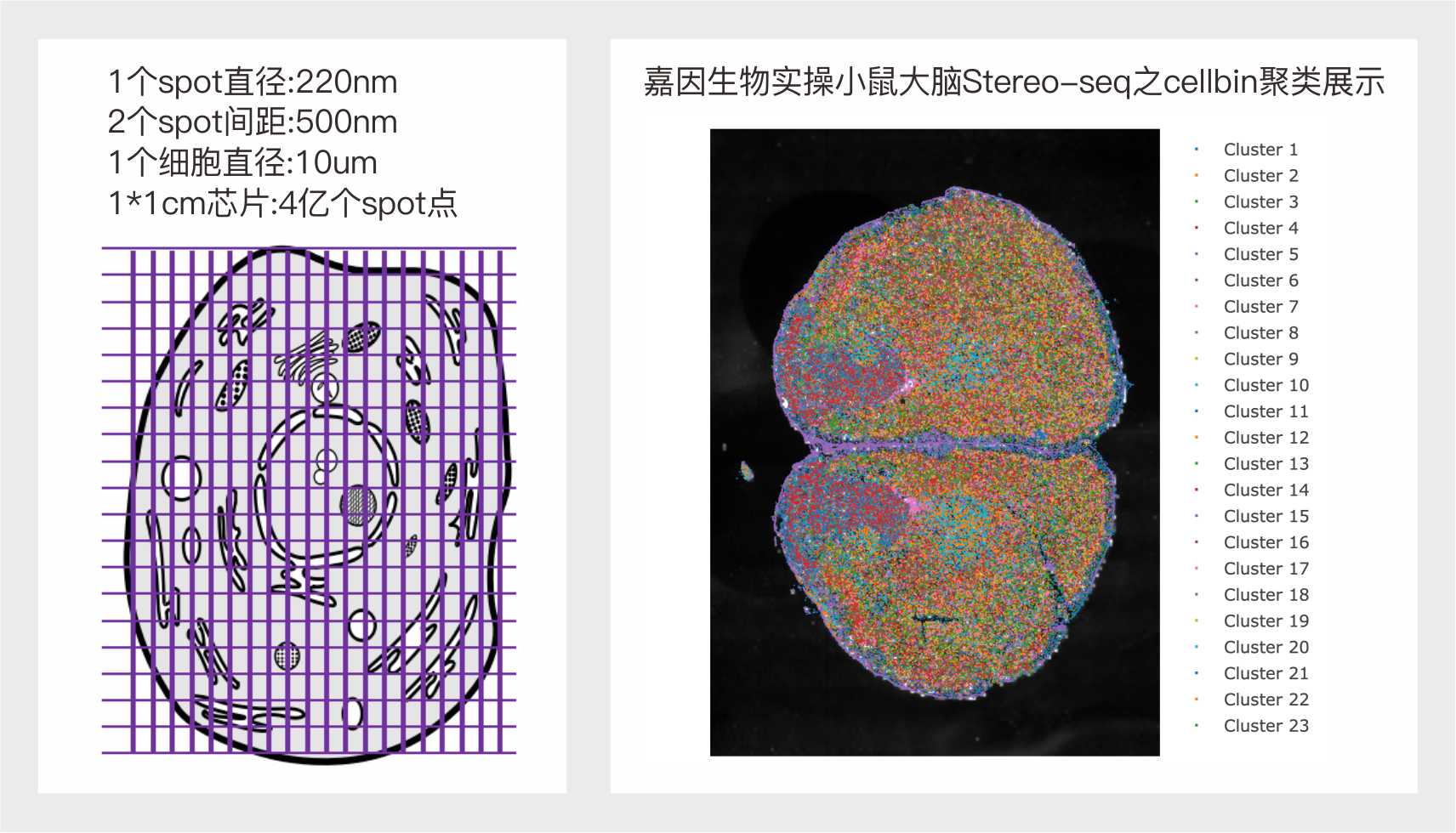
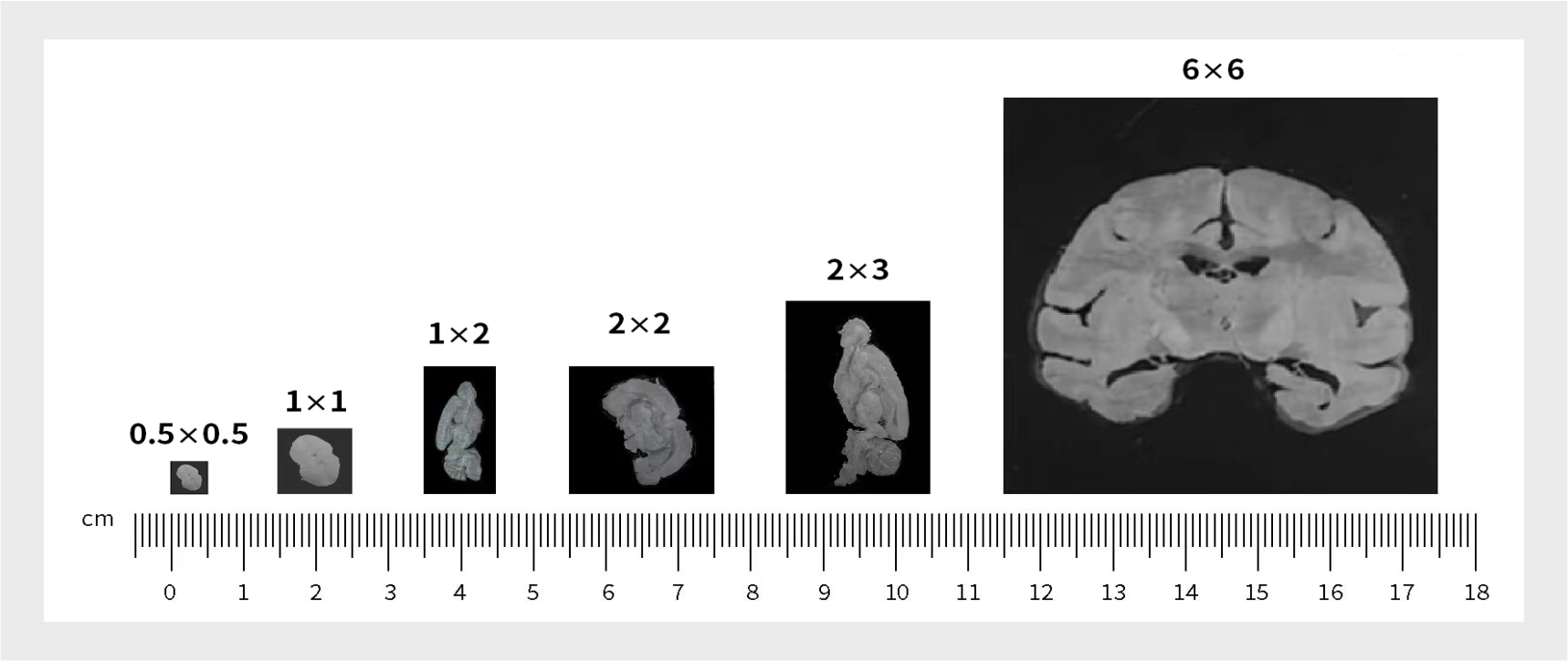
优势二: 厘米级全景视场,检测组织大小可定制。(目前商用芯片规格:1cm*1cm;0.5cm*0.5cm)
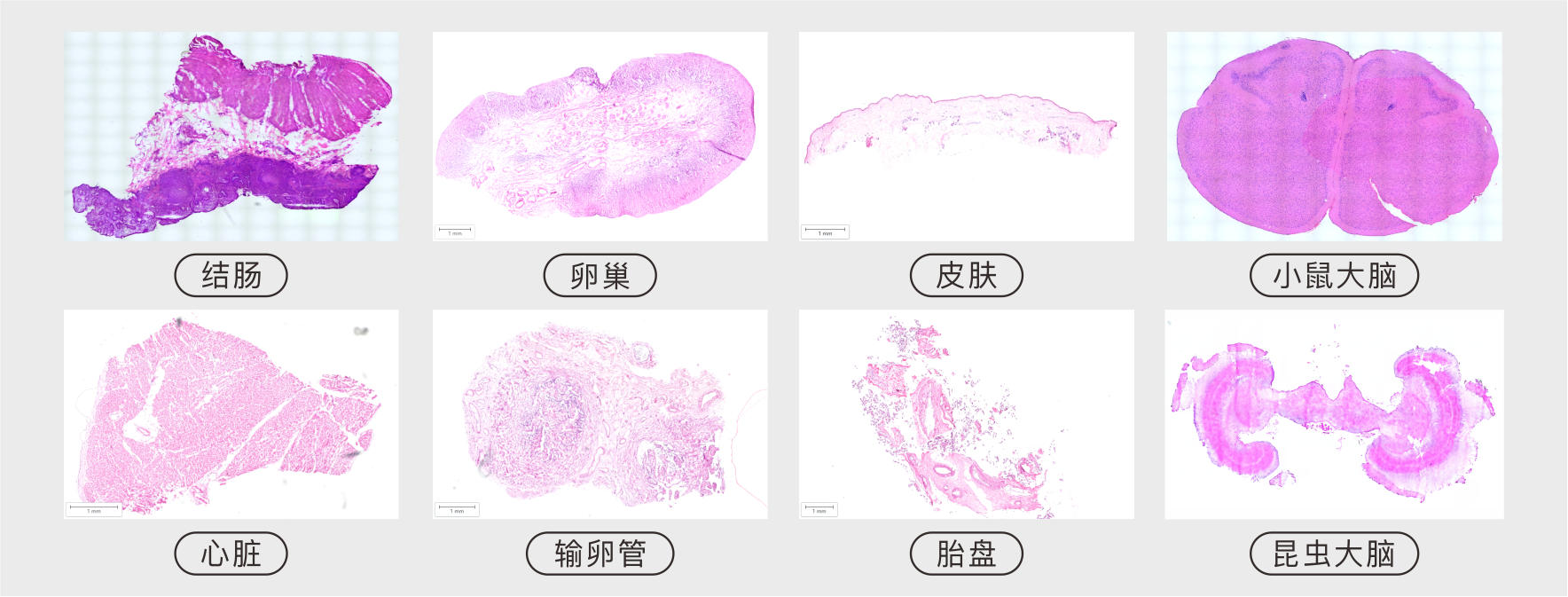
优势三: 物种组织不设限,人、动物及植物多覆盖。


▍嘉因生物Stereo-seq服务优势
优势一:嘉因生物切片事业部核心成员拥有8年临床病理学经验。专业攻克各种器官类型切片。
优势二:自2023年1月起,历经9个月时间,嘉因生物已搭建好Stereo-seq分析流程。各类空转定制化分析需求应有尽有,满足老师不同研究需求。
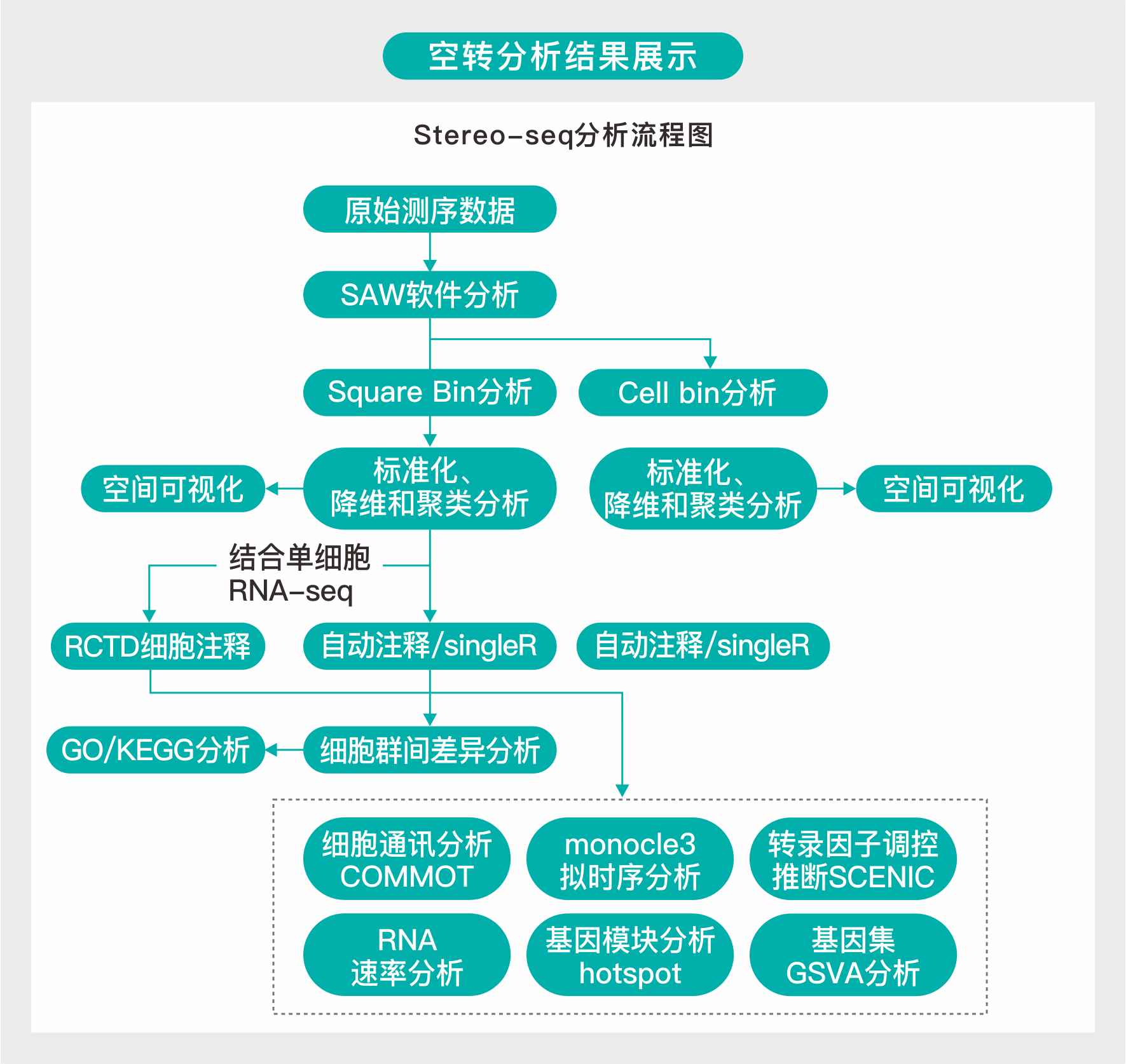
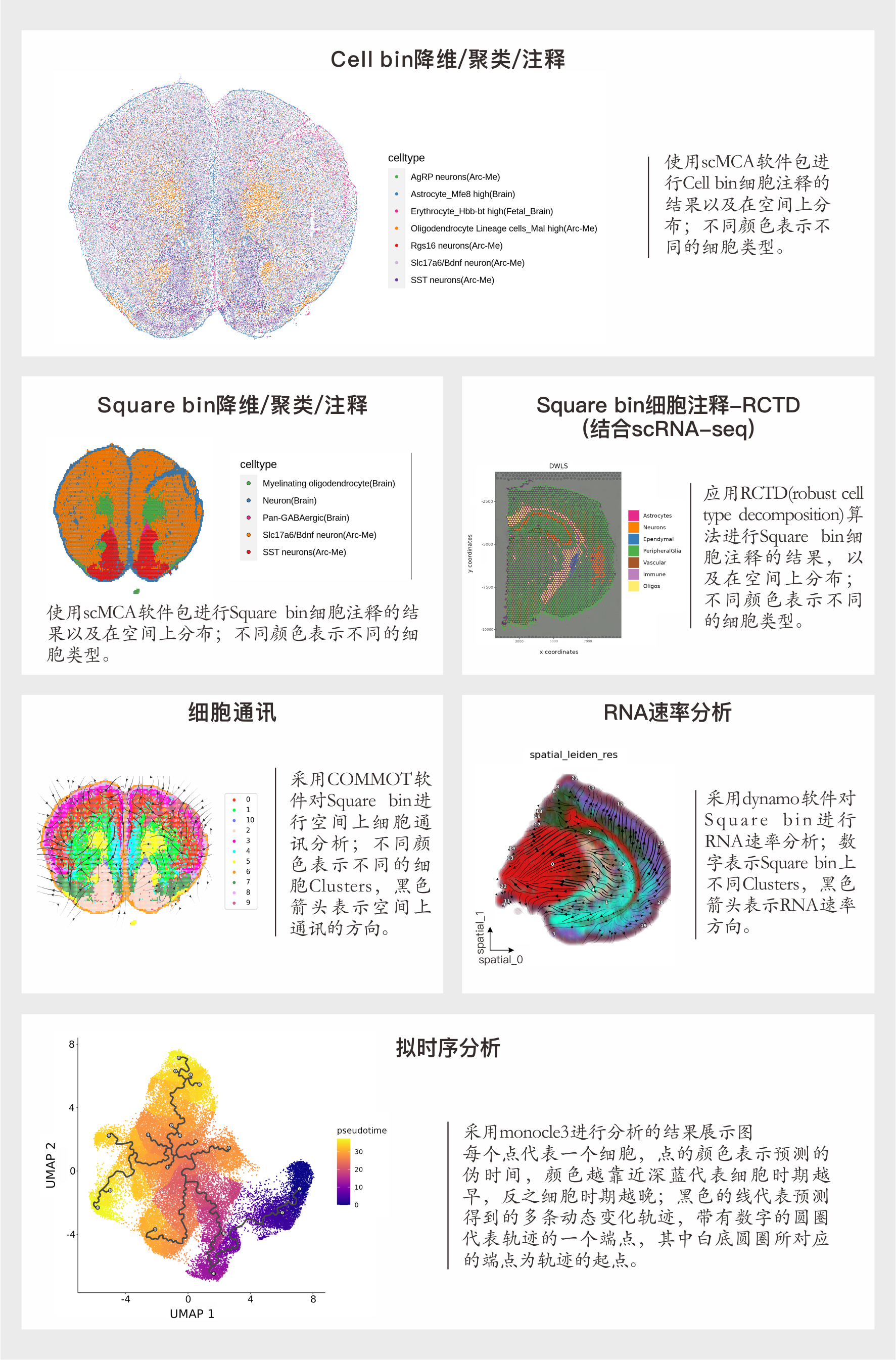
原始数据在下机后,首先使用SAW软件进行分析,得到Square bin表达矩阵和Cell bin表达矩阵。
在Square bin中,每个bin表示一个固定大小的区域,也是时空芯片上每个DNB表达热图的一个像素点。bin1是最小的分析单元,而bin50表示由50*50=2500个DNB区域组成的数据。Cell bin是根据ssDNA染色分割的单个细胞的区域数据单位。
对Cell bin进行以下分析步骤:
(1) 进行数据标准化、降维和聚类,并进行空间可视化分析。
(2) 使用SingleR进行自动细胞注释。
对Square bin进行以下分析步骤:
(1) 进行数据标准化、降维和聚类,并进行空间可视化分析。
(2) 使用SingleR进行自动细胞注释。如果有配套的scRNA-seq数据则还会进行RCTD细胞注释。
(3) 进行细胞群差异分析和GO/KEGG分析。
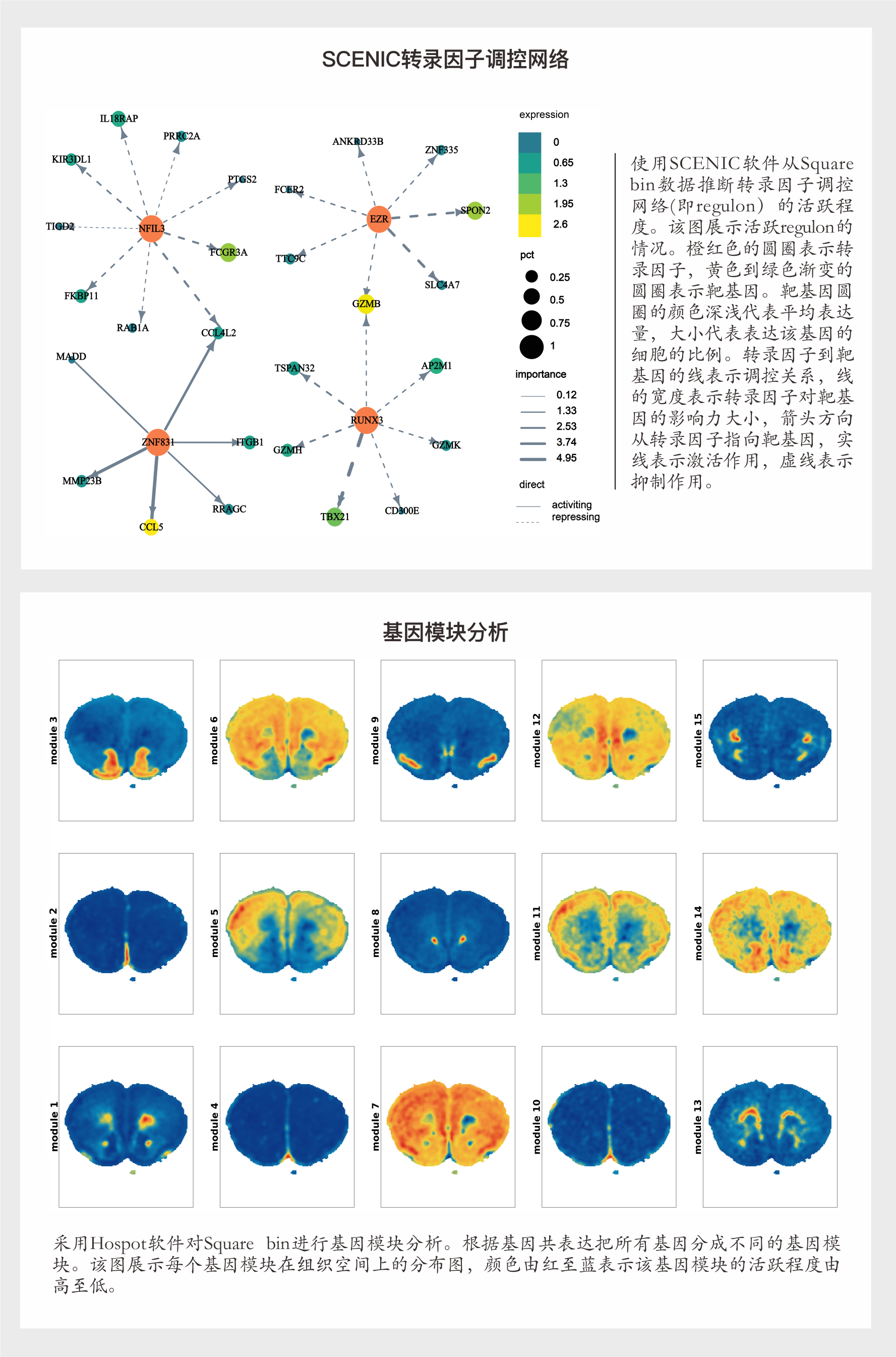
(4) 根据细胞注释的分群,进行细胞通讯分析、拟时序分析、转录因子调控推断、RNA速率、基因模块分析和基因集GSVA富集分析。
▍客户文章精选
| 样本类型 | 文章标题 | 杂志 | 影响因子 |
| / | A cellular hierarchy in melanoma uncouples growth and metastasis | Nature | 69.504 |
| 小鼠嗅球、脑、胚胎 | Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays | Cell | 66.85 |
| 食蟹猴脑 | Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex | Cell | 66.85 |
| 墨西哥蝾螈脑 | Single-cell Stereo-seq Reveals Induced Progenitor Cells Involved in Axolotl Brain Regeneration | Science | 56.9 |
| 人肝 | An invasive zone in human liver cancer identified by Stereo-seq promotes hepatocyte-tumor cell crosstalk, local immunosuppression and tumor progression | Cell research | 46.351 |
| / | Unveiling the hidden battlefield: dissecting the invasive zone in liver cancer | Cell research | 46.351 |
| / | Clinical and translational values of spatial transcriptomics. | Signal transduction and targeted therapy | 38.12 |
| 人胚胎 | A single-cell transcriptome atlas profiles early organogenesis in human embryos. | Nature cell biology | 28.213 |
| 猴子腹直肌 | Implantation underneath the abdominal anterior rectus sheath enables effective and functional engraftment of stem-cell-derived islets | Nature Metabolism | 19.89 |
| / | Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. | Nature communications | 17.69 |
| 食蟹猴脑 | Spatially resolved gene regulatory and disease-related vulnerability map of the adult Macaque cortex | Nature communications | 17.69 |
| / | Spatial-ID: a cell typing method for spatially resolved transcriptomics via transfer learning and spatial embedding. | Nature communications | 17.69 |
| / | Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST | Nature communications | 17.69 |
| / | Single-Nucleus RNA Sequencing and Spatial Transcriptomics Reveal the Immunological Microenvironment of Cervical Squamous Cell Carcinoma | Advanced Science | 17.521 |
| 大豆节结 | Integrated single-nucleus and spatial transcriptomics captures transitional states in soybean nodule maturation. | Nature plants | 17.352 |
| 斑马鱼胚胎 | Spatiotemporal mapping of gene expression landscapes and developmental trajectories during zebrafish embryogenesis | Developmental Cell | 13.417 |
| 果蝇胚胎 | High-resolution 3D spatiotemporal transcriptomic maps of developing Drosophila embryos and larvae | Developmental Cell | 13.417 |
| 拟南芥叶子 | The single-cell stereo-seq reveals region-specific cell subtypes and transcriptome profiling in Arabidopsis leaves | Developmental Cell | 13.417 |
| / | Spatial transcriptome analysis on peanut tissues shed light on cell heterogeneity of the peg. | Plant biotechnology journal | 13.263 |
| 小鼠脑 | A Cellular Resolution Spatial Transcriptomic Landscape of the Medial Structures in Postnatal Mouse Brain. | Frontiers in cell and developmental biology | 6.081 |
| 人胎盘 | Resolving the gene expression maps of human first-trimester chorionic villi with spatial transcriptome | Frontiers in cell and developmental biology | 6.081 |
| 小鼠脑 | Developmental deficits of MGE-derived interneurons in the Cntnap2 knockout mouse model of autism spectrum disorder | Frontiers in cell and developmental biology | 6.081 |
| / | Spatially resolved transcriptomics: a comprehensive review of their technological advances, applications, and challenges | Journal of Genetics and Genomics | 5.9 |
| / | VT3D: a visualization toolbox for 3D transcriptomic data | Journal of Genetics and Genomics | 5.9 |
| 小鼠脾脏 | Microbiota-mediated shaping of mouse spleen structure and immune function characterized by scRNA-seq and stereo-seq | Journal of Genetics and Genomics | 5.9 |
